
- Home
- About Us
- Service
One-stop Solutions
One-stop Solutions
IVD Products
One-stop Solutions
One-stop Solutions
IVD Products
One-stop Solutions
One-stop Solutions
IVD Products
- Application
- Technology
- Resource
- Contact Us

- Home
- About Us
- Service
- Clinical Products +
- Multi-omics Tech Services +
- Biopharma Services
- Application
- Technology
- Resource
- Contact Us

LIUDUS®
HaploX's proprietary molecular technologies cover all the major steps within the wet lab NGS workflow, optimizing each step to ensure superior nucleic acid isolation and sequencing library preparation while significantly reducing sequencing errors. This comprehensive approach yields high-quality results with maximized efficiency.
01
CUBE-ctDNA
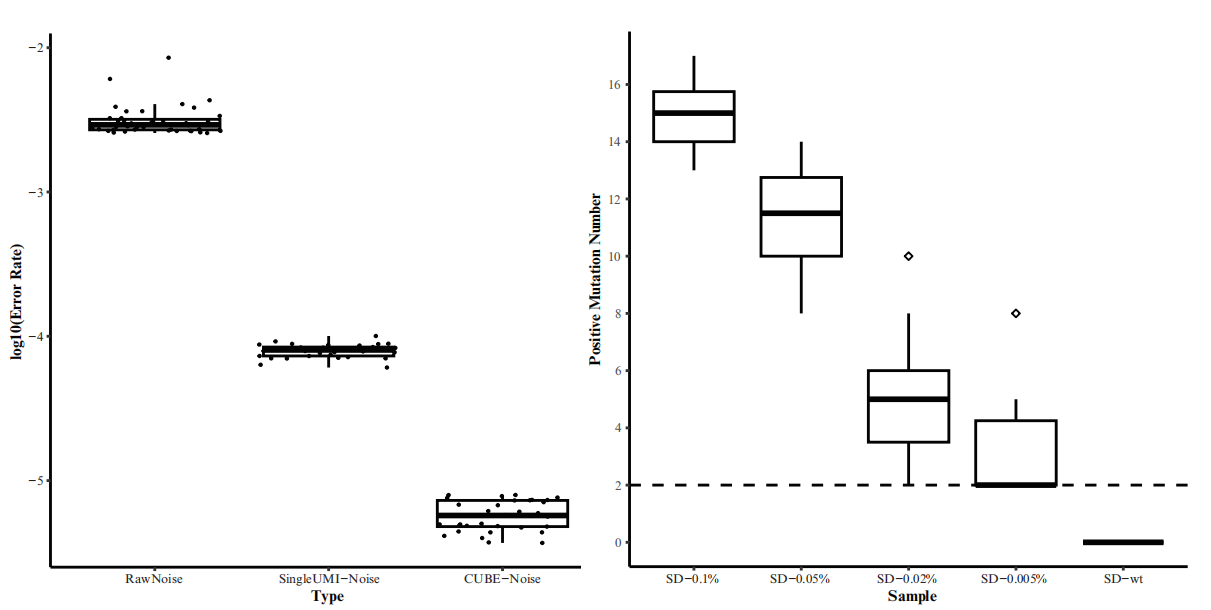
CUBE-ctDNA is HaploX's innovative single-molecule barcoding technology designed to enhance the signal-to-noise ratio in ultra-deep sequencing. During library preparation, this technology assigns two unique molecular identifiers (UMIs) to both ends of each double-stranded DNA molecule. This allows each sequence read, post-PCR amplification and sequencing, to be traced back to the original DNA molecule. True mutations appear consistently in reads tagged with the same UMIs, while false mutations, such as PCR and sequencing errors, appear only in a fraction of these reads. By attaching UMIs to both DNA strands, CUBE-ctDNA leverages sequence complementarity to filter out false mutations detected in only one strand.
Through CUBE-ctDNA technology, we significantly reduce NGS background noise from an industry standard of 0.1% to 0.00058% in preclinical validation studies, allowing ultra-deep sequencing with a ctDNA LoD as low as 0.005%.
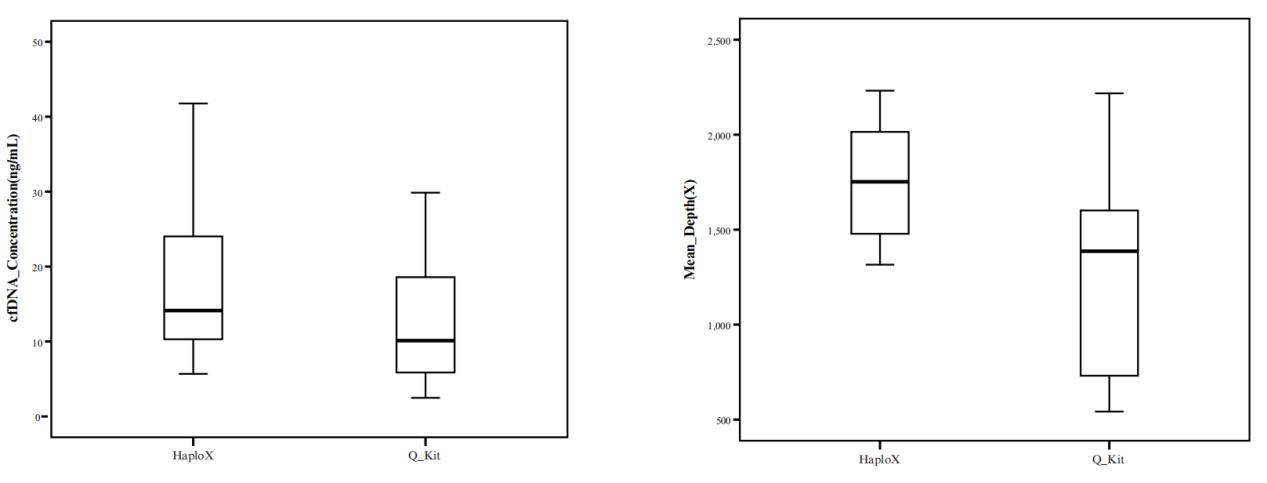
Compared to a competitive international DNA extraction product in our preclinical validation studies, Effentration® consistently achieved higher cfDNA isolation efficiency and greater average sequencing depth.
02
Effentration®
DNA extraction is a critical first step to the success of NGS-based variant detection. To that end, HaploX developed Effentration® for effective cfDNA extraction. Via the use of lysis, binding and washing solutions with proprietary formulations, Effentration® can effectively enrich low-abundance cfDNA and remove undesirable nucleic acid contaminants.
03
HAPCap®
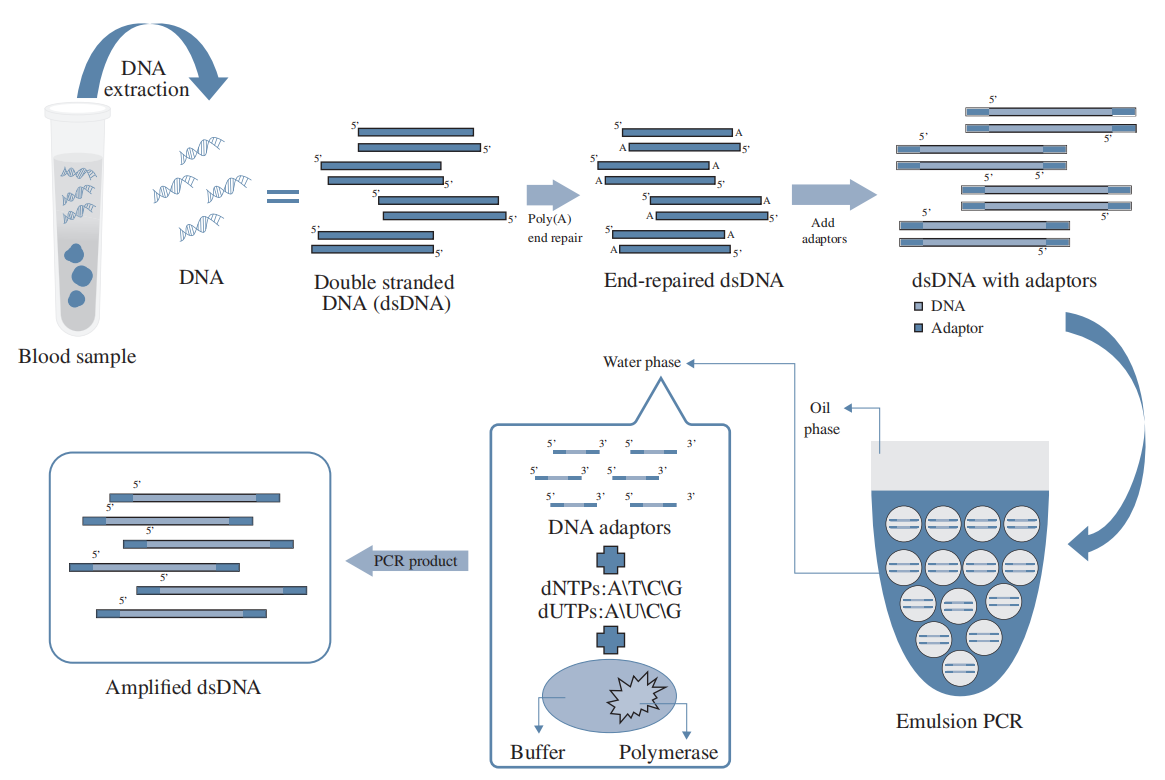
During library preparation, the initial DNA material from biopsy samples needs PCR amplification to ensure sufficient DNA for sequencing. Since ctDNA is a minor fraction of cfDNA, conventional PCR amplification dilutes detection probability by favoring dominant cfDNA types. To tackle this, HaploX developed HAPCap® for ctDNA enrichment during DNA capture. This process begins with emulsion PCR, where input DNA molecules are amplified in separated micro-volume water-in-oil droplets. Then, ctDNA-specific, biotin-labeled DNA probes hybridize to ctDNA, which is captured using streptavidin magnetic beads. HAPCap® effectively removes PCR amplification bias, enabling selective amplification of low-abundance ctDNA to enhance mutation detection sensitivity.
HAPCap® Technical Flow Chart
HaploX has pioneered the development of cutting-edge bioinformatics software, machine-learning models, and data libraries. These tools form the backbone of our data processing infrastructure, enabling efficient sequencing data analysis, error removal, and precise detection of low-frequency mutations.
Bioinformatics Software
01
Fastp
HaploX's Fastp enhances data preprocessing by integrating features from commonly used tools for quality control, adapter trimming, and per-read quality pruning. Developed in C++ with multi-threading support, Fastp operates significantly faster, as validated in studies showing it to be two to five times quicker while maintaining or surpassing the quality of data filtering and mutation detection.
Published in Bioinformatics in September 2018, Fastp is available as an open-source tool on GitHub.
02
Gencore
HaploX’s Gencore efficiently removes redundant sequencing information and offers memory-efficient PCR duplicate removal and consensus read generation for NGS data, with or without UMIs. It provides informative statistical reports to aid quality control and downstream analysis. Published in BMC Bioinformatics in December 2019, Gencore is available as an open-source tool on GitHub.
03
MutScan
MutScan is a high-performance tool for detecting and visualizing target mutations, significantly improving detection sensitivity. Unlike traditional pipelines, MutScan can directly detect target mutations from raw FASTQ files using a high-error-tolerance string searching algorithm. It validates detected mutations and those identified by conventional pipelines through an HTML report, evaluating mutation confidence via multiple metrics.
Published in BMC Bioinformatics in January 2018, MutScan is available as an open-source tool on GitHub.
04
GeneFuse
Designed for clinical applications, GeneFuse offers fast, sensitive detection and visualization of target gene fusions. It bypasses the alignment step typical of most tools, scanning raw sequencing data directly, thereby achieving higher sensitivity and specificity.
Published in the International Journal of Biological Sciences in May 2018, GeneFuse is available as an open-source tool on GitHub.
05
FineMSI
FineMSI is HaploX's patented tool for analyzing microsatellite loci and determining MSI status using the earth movers' distance (EMD) method. It evaluates the dissimilarity between MSI-high and MSI-low data distributions, offering greater sensitivity and specificity than standard NGS-based MSI methods. FineMSI showcases its potential as a precise method for MSI status determination.
Machine Learning
HaploX leverages machine learning to tackle complex image denoising and target recognition challenges. By training workflows on extensive real-world data, we can minimize manual intervention, enhancing our ability to detect low-frequency cancer mutations and thereby improving the sensitivity and specificity of genetic tests.
01
MrBam
MrBam, HaploX's patented software, uses machine learning models trained on vast datasets of background noise and false-positive mutation sites to filter variant noise effectively.
02
TCRnodseek
TCRnodseek accurately classifies benign and malignant pulmonary nodules using a support vector machine that integrates TCR characteristics and clinical information. Based on a study of 99 individuals with indeterminate lung nodules, TCRnodseek demonstrated robust sensitivity (76%), specificity (91%), accuracy (84%), and an AUC of 0.8. The clinical results were published in Signal Transduction and Targeted Therapy in October 2022.
Luo, H., Zu, R., Huang, Z. et al. Characteristics and significance of peripheral blood T-cell receptor repertoire features in patients with indeterminate lung nodules. Sig Transduct Target Ther 7, 348 (2022). https://doi.org/10.1038/s41392-022-01169-7
Data Libraries

01
Data Libraries
HaploX's proprietary data libraries, built from extensive real-world sequencing data, facilitate mutation interpretation:
1. HapKnow: Annotates over 1.4 million tumor somatic mutations.
2. HapHeal: Annotates over 1.3 million hereditary mutations.
These libraries underpin our HapReport system, providing automated mutation reporting with concise clinical significance interpretations.
By integrating innovative bioinformatics tools, machine learning models, and comprehensive data libraries, HaploX continues to lead in genetic testing solutions, ensuring precise and efficient mutation detection and interpretation.
HaploX has meticulously developed an end-to-end IT infrastructure that forms the backbone of our digital, automated, and smart laboratory operations.
01
HapLab®
Comprehensive Laboratory Information Management System
HapLab® integrates HaploX’s proprietary molecular technologies and in-house bioinformatics tools under the LIUDUS®. This LIMS system ensures seamless, information-based, closed-loop management of our entire NGS workflow. HapLab® consists of four core modules:
1. Order Management Module:
• Manages systematic accession and tracking of clinical samples.
• Ensures sample integrity and proper chain of custody.
2. Wet Lab Workflow Management Module:
• Automates project resourcing and organization of experimental workflows.
• Reduces user intervention, increases wet lab efficiency, and maintains data integrity.
3. Bioinformatics Analysis Module:
• Executes complex multi-step bioinformatics workflows with automatic parallel analysis.
• Enhances analysis efficiency.
4. Report Interpretation Module:
• Integrates multidimensional patient information.
• Facilitates data management and mining at both patient and population levels.
02
HapYun®
Advanced Cloud Platform for Data Management
HapYun® is our proprietary one-stop cloud platform designed for efficient and scalable data management, storage, analysis, and delivery. Key features of HapYun® include:
• A rich library of modular, out-of-the-box pipelines and tools for various molecular diagnostics applications.
• Efficient, customizable analysis of high-throughput genomics data at scale.
• Automatic task execution, real-time operation logs, and performance monitoring.
By embedding these advanced systems, HaploX ensures superior efficiency, accuracy, and scalability in genetic testing, reinforcing our commitment to cutting-edge molecular diagnostics.
Address
Units 610, 6/F, Building 16W, Phase 3, HongKong Science Park, New Territories, HongKong
Phone
+852 69410851
customer_service@haplox.net
© 2025 HaploX Ltd. All Rights Reserved.