
- Home
- About Us
- Service
- Application
- Technology
- Resource
- Contact Us

Eukaryotic mRNA Sequencing
mRNA sequencing is an advanced gene expression analysis technique that uses high-throughput sequencing to quantitatively analyze mRNA in cells. It identifies known and novel transcripts, and reveals gene expression levels and alternative splicing. Widely used in biological research, disease studies, drug development, and diagnostics, it offers critical insights into gene regulation and function.
Work flow
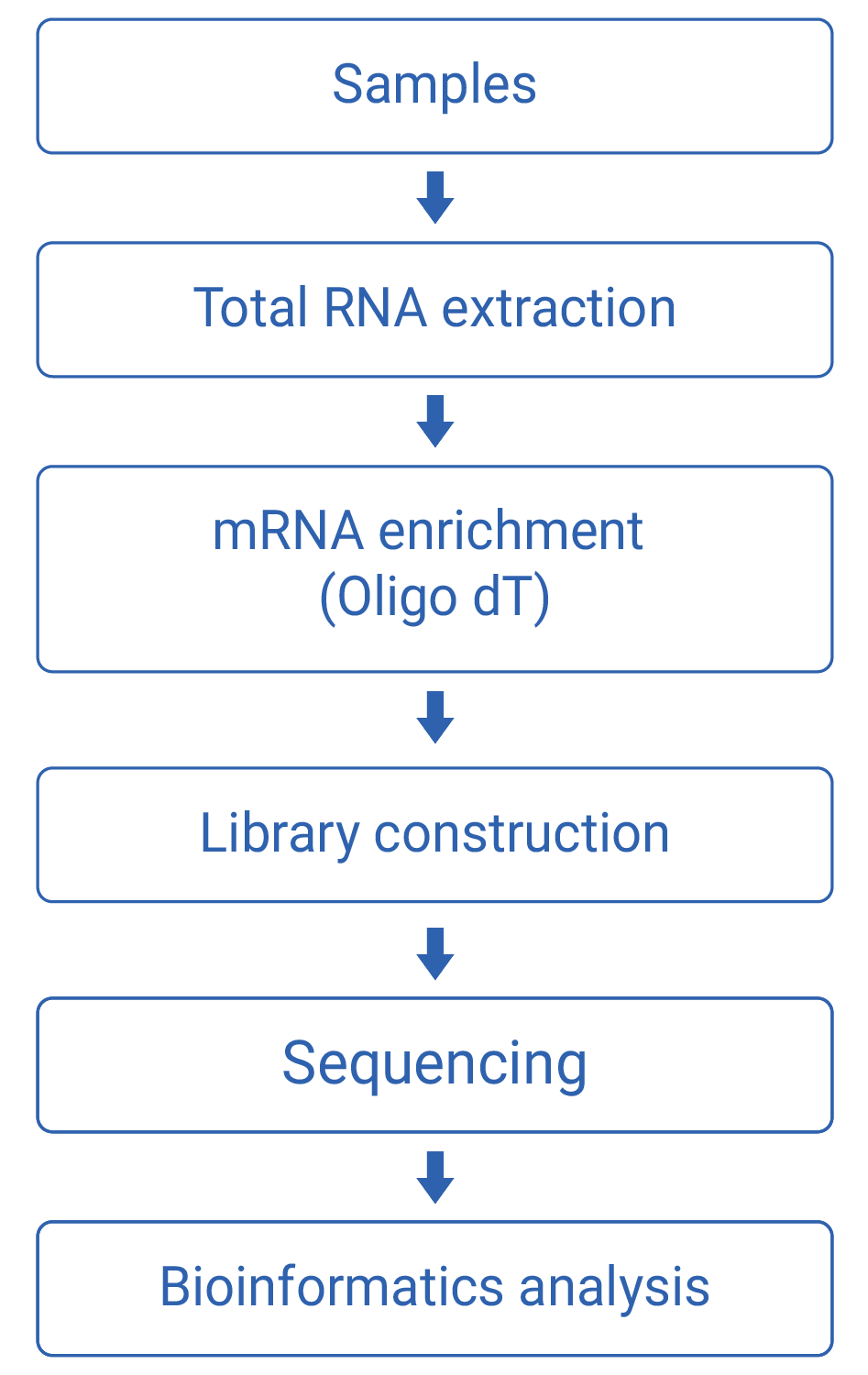
Technical Parameters
| Sequencing range | mRNA |
| Sequencing strategy | NGS PE150 |
| Sequencing throughput | 6Gb |
| Data quality | Fastq files, Q30≥85% |
| Data analysis | Standard+advanced analysis |
| TAT | Standard: 25 WD |
Applications
| Gene Expression Analysis | Discovery of Novel Transcripts and Splice | Drug Mechanism of Action Studies |
| Disease Diagnosis and Biomarker Discovery | Regulatory Network Analysis | Cancer Research and Treatment |
Bioinformatics Analysis
1.Data Quality Control: Remove adapter contamination and low-quality reads from the raw data.
2.Reference Genome Alignment: Statistics of alignment results, display of alignment region distribution.
3.Gene Expression Analysis: Reads count statistics, calculation of expression levels (FPKM), analysis of sample correlations.
4.Differential Gene Expression Analysis: Statistics of differentially expressed genes, volcano plots, cluster heatmaps, Venn diagrams.
5.Differential Gene Functional Enrichment Analysis: GO (Gene Ontology) significance enrichment analysis, KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis.
6.Differential Gene Protein Interaction Network Analysis.
7.Differential Alternative Splicing Analysis and Statistics.
8.Fusion Gene Analysis: Detection and annotation of fusion genes, display of chromosomal distribution of fusion genes.
9.Variant Detection and Statistics.
Advantages
![]() No specific probe required
No specific probe required
![]() No species genome information required
No species genome information required
![]() Detect known transcripts and discover unknown transcripts
Detect known transcripts and discover unknown transcripts
![]() Advanced analysis & customized analysis provided
Advanced analysis & customized analysis provided
