
- Home
- About Us
- Service
- Application
- Technology
- Resource
- Contact Us

Astral-InDepth Blood Proteomics
Product Overview
Blood, being an easily accessible, stable, and minimally invasive biological sample, is regarded as an ideal source for biomarker evaluation. However, the presence of highabundance proteins in blood greatly hampers the depth of proteomic identification.
Leveraging the Thermo Orbitrap™ Astral™ high-resolution mass spectrometer, HaploX's InDepth Blood Proteomics service overcomes challenges posed by high-abundance proteins in blood. It combines nanomagnetic bead-based low-abundance protein enrichment with Astral-DIA full-scan acquisition to enable high-throughput, deep-coverage, and sensitive blood proteomic analysis. Unprecedented speed, coverage, sensitivity, and quantitative accuracy make it ideal for trace protein detection and large-scale studies, facilitating in-depth blood biomarker discovery.
Advantages
![]() Minimal Sample Requirement: Only 50 μL starting volume required.
Minimal Sample Requirement: Only 50 μL starting volume required.
![]() Superior Identification Depth: Over 5-fold more proteins identified vs. conventional methods (>5,000 total), with outstanding enrichment of low-to-medium abundance proteins.
Superior Identification Depth: Over 5-fold more proteins identified vs. conventional methods (>5,000 total), with outstanding enrichment of low-to-medium abundance proteins.
![]() Customizable workflows: Standard 8/24-minute detection, plus personalized solutions.
Customizable workflows: Standard 8/24-minute detection, plus personalized solutions.
![]() Full-process QC: Comprehensive system covering sample prep and MS analysis, backed by large-cohort study experience.
Full-process QC: Comprehensive system covering sample prep and MS analysis, backed by large-cohort study experience.
![]() Enhanced Depth: Disease-derived plasma samples yield more low-abundance proteins, aiding biomarker discovery.
Enhanced Depth: Disease-derived plasma samples yield more low-abundance proteins, aiding biomarker discovery.
![]() Stable Performance: Automation + full QC system reduces cohort detection risks.
Stable Performance: Automation + full QC system reduces cohort detection risks.
Product Parameters
| Indicator | Parameter |
|---|---|
| Detection Range | Proteins in serum/plasma |
| Instrument Platform | Thermo Orbitrap Astral |
| Turnaround Time | 30 Working Days |
| Delivery Standard | Raw Data + Standard Analysis |
Sample Requirements
| Sample Type | Samples | Sample Size |
|---|---|---|
| Liquid Samples | Plasma/serum | 50uL |
Workflow
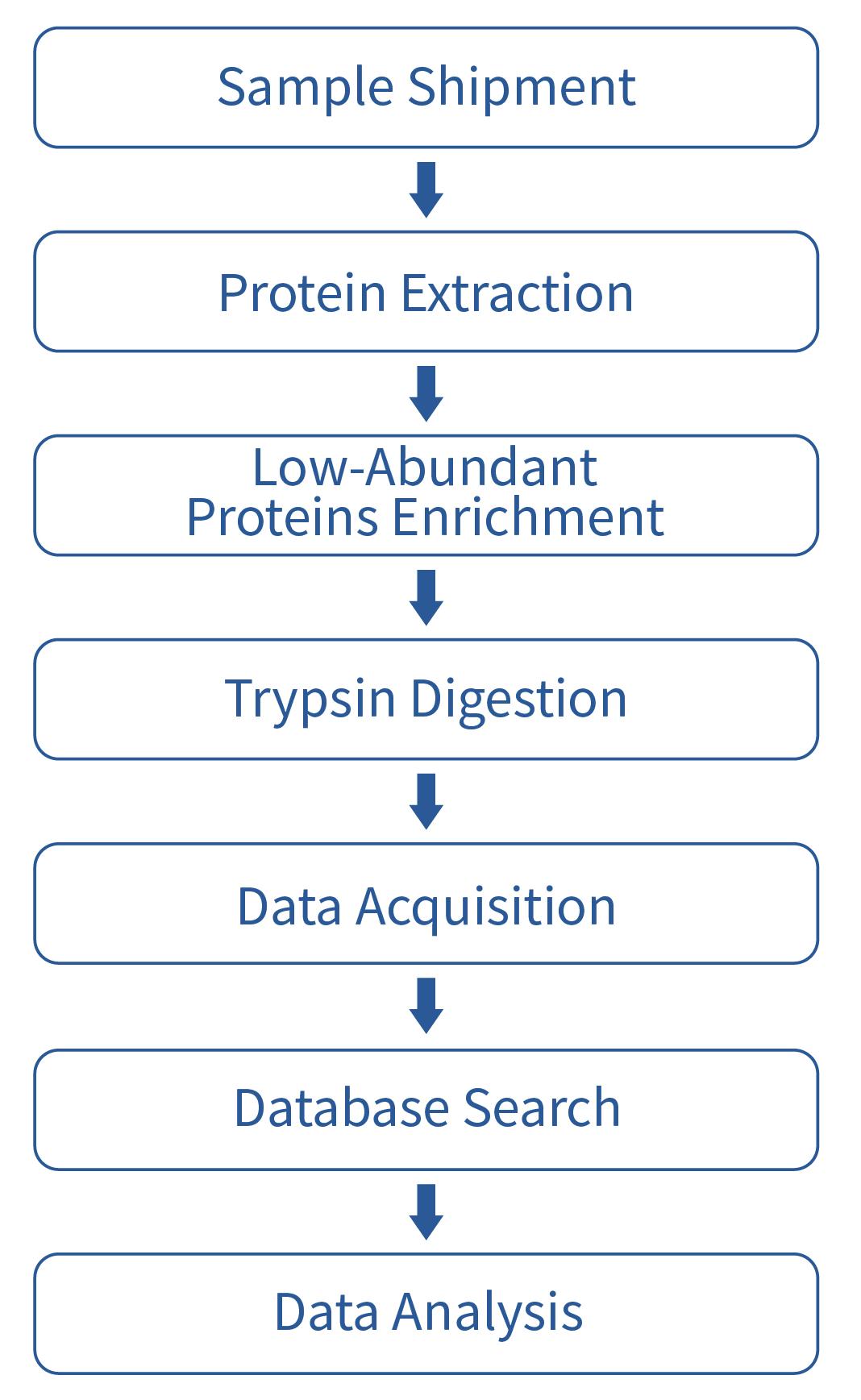
Analysis Contents
| Standard Data Analysis | |
|---|---|
| Quality Control | Peptide charge/length/FDR distribution, missed cleavage rate statistics. |
| Identification | Statistical results of identified proteins for individual samples and the overall dataset. |
| Functional Analysis | Pathway enrichment (GO, KEGG, Reactome, Wikipathways), chromosomal localization, subcellular localization. |
| Differential Expression | Statistics of differentially expressed proteins, bar plots, volcano plots, cluster analysis, pathway enrichment of differentially expressed proteins. |
| Multi-group Comparison | Statistics of differentially expressed proteins and functional analysis |



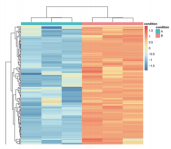
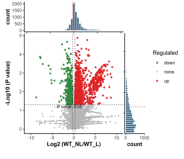
Application Scenarios
![]() Biomarker Discovery
Biomarker Discovery
![]() Disease Mechanism Investigation
Disease Mechanism Investigation
![]() Therapeutic Evaluation
Therapeutic Evaluation
![]() Drug R&D and Personalized Medicine
Drug R&D and Personalized Medicine
![]() Disease Diagnosis and Prevention
Disease Diagnosis and Prevention
FAQ
Q:Why are specialized proteomic workflows necessary for blood samples such as serum and plasma?
A: Serum and plasma pose unique analytical challenges in proteomics due to the presence of highly abundant proteins (e.g., albumin, immunoglobulins), which account for >90% of total protein. These proteins interfere with the detection of low-abundance species, restricting standard workflows to identifying only several hundred proteins. Thus, specialized blood proteomics methods are critical to achieve comprehensive protein coverage and facilitate the discovery of disease-related biomarkers.
