
- Home
- About Us
- Service
- Application
- Technology
- Resource
- Contact Us

10X Single Cell Transciptome
Product Overview
The 10X single-cell technology, based on microfluidics and droplet technology, uses an oil-in-water system to encapsulate cells with barcoded gel beads into Gel Beads in Emulsion (GEMs) for separation and labeling. It constructs high-throughput sequencing libraries and is widely used in studying cell heterogeneity, analyzing tumor microenvi-ronments, and detecting immune cell populations.
Advantages
(1) Experience in dissociating tissue samples of over 200 tumor types
(2) Multi-omics joint analysis solution
(3) Over 10 unique bioinformatics algorithms
(4) Clinical-grade experimental management
Product Parameters
| Indicator | Parameter |
|---|---|
| Detection Region | 3' mRNA |
| Instrument Platform | 10X Genomics |
| Sequencing Data Volume | 30K~80K reads/Cell |
| Turnaround Time | 35 working days without analysis; 55 working days with standard analysis |
| Applicable Species | Humans, mice, rats & other eukaryotes |
| Number of captured cells | 500~15,000 |
| Applicable Sample Type | Cell suspension, fresh tissue, fresh blood, peripheral blood mononuclear cells (PBMCs), puncture sample and cryopreserved tissue |
Sample Requirements
| Sample Type | Recommended Sample Quantity | Transport Conditions | Shipping Time |
|---|---|---|---|
| Puncture Sample | ≥2 pieces (16G puncture needle, length >1.5cm; 3–4 pieces for breast cancer puncture / needle outer diameter ≥1.2mm) | In tissue preservation solution, shipped with ice packs or crushed ice at 2–8℃ | 24–48h |
| Fresh Blood (default for PBMC extraction) | 2–4mL, minimum 1mL | Collected in EDTA blood collection tubes, shipped with crushed ice or crushed ice at 2–8℃ | 4–12h |
| Fresh Tissue | ≥200mg (size of a soybean, 0.5cm³) | In tissue preservation solution, shipped with ice packs or crushed ice at 2–8℃ | 24–72h |
| Cryopreserved Tissue | ≥200mg (size of a soybean, 0.5cm³) | Quick-frozen in liquid nitrogen and shipped with dry ice | Unlimited |
| Cryopreserved PBMC | No less than 300,000 cells (minimum 1mL) | Shipped with dry ice after gradient cooling cryopreservation | Unlimited |
| Single-cell Suspension | Cell count≥3*105 Cell viability > 90%,clumping rate <20% | Shipped with dry ice after gradient cooling cryopreservation | Fresh cell suspension:within 4h |
| Body Fluid | Total cell count ≥105(8–15mL ascites, joint fluid, cerebrospinal fluid, etc., minimum 4mL) | Shipped with crushed ice; ensure buffer protection during transportation to avoid severe shaking of samples | Fresh body fluid:within 4h |
| Biopsy sample | ≥2 strips | 2-8℃ | Ship in preservative within 48h |
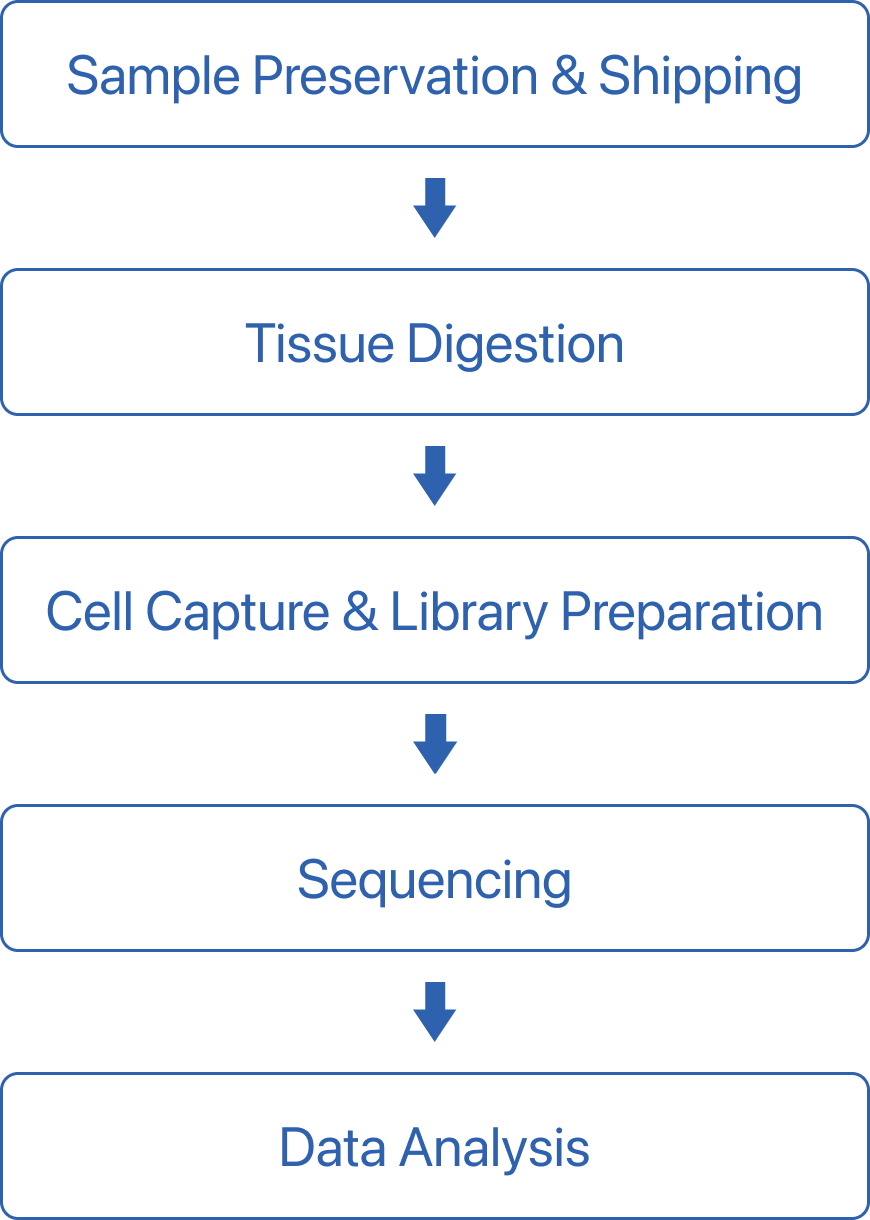
Workflow
Analysis Contents
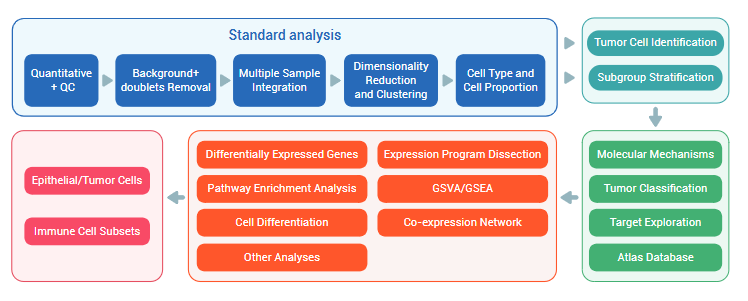
| Analysis Type | Analysis Content |
|---|---|
| Standard Analysis |
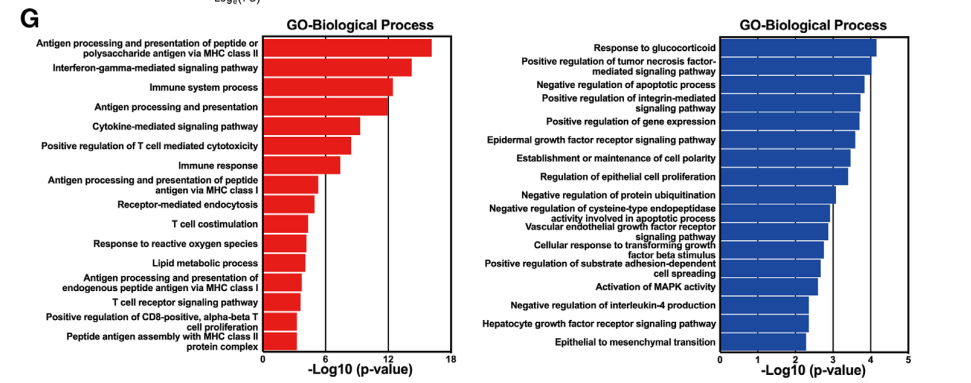
1. Raw data quality control 2. Cell number estimation 3. Genome alignment 4. Expression level statistics 5. Cell filtering 6. Data normalization 7. Cell type annotation 8. Cell count statistics 9. Marker gene identification 10. Screening of highly expressed genes in clusters 11. Functional analysis (GO Analysis) 12. Pathway analysis (Pathway Analysis). |
| Analysis Type | Analysis Content |
|---|---|
| Advanced Analysis |
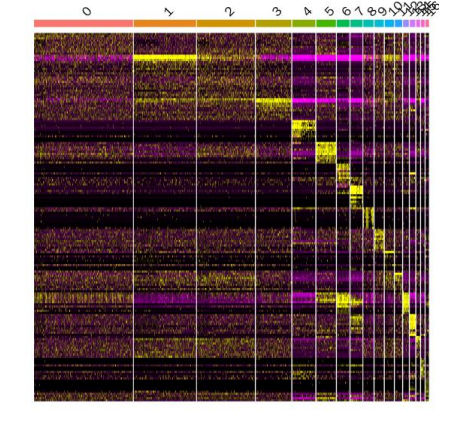
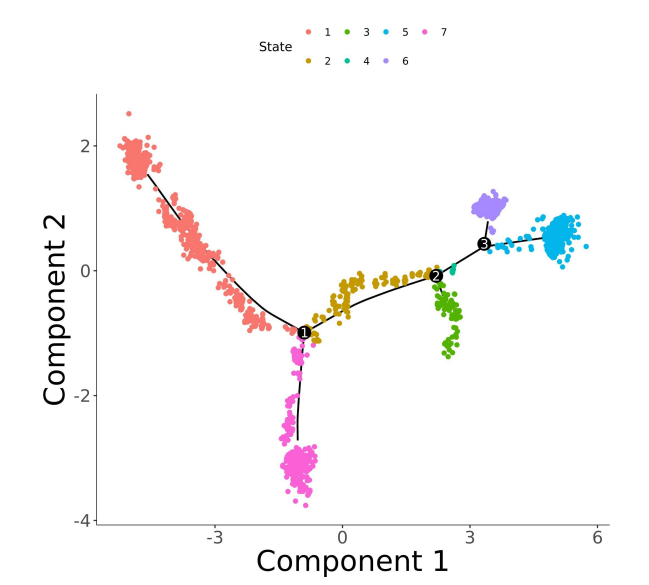
1. Subpopulation re-clustering 2. Inter-group differential gene analysis 3. GO/KEGG enrichment analysis 4. GSVA and differential pathway analysis 5. GSEA 6. Cell-cell interaction analysis 7. inferCNV/CopyKat analysis 8. Subpopulation expression signature decomposition (cNMF) 9. Monocle2/Monocle3 pseudotime analysis 10. RNA velocity analysis 11. CytoTRACE/cell stemness analysis 12. Transcription factor regulatory activity and network analysis |
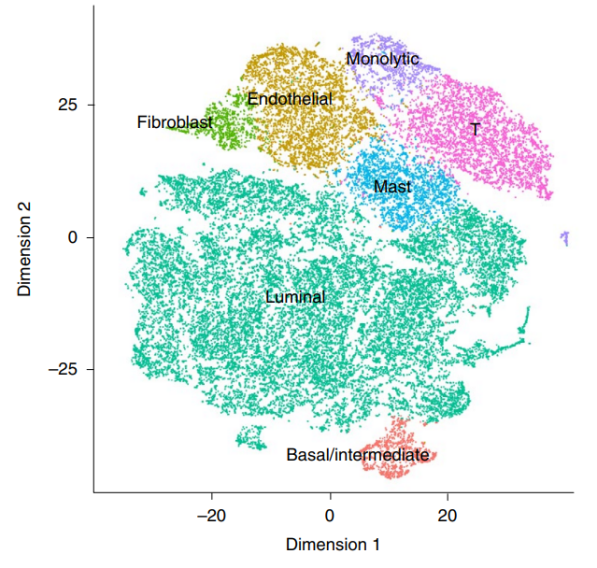
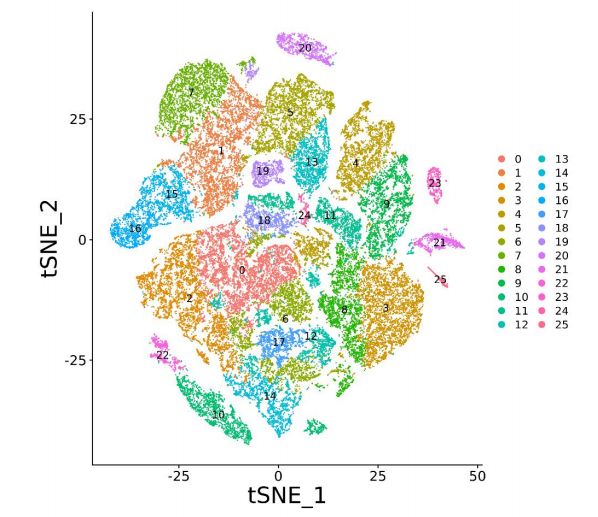
Application Scenarios
![]() Tumor Microenvironment Research
Tumor Microenvironment Research
![]() Developmental Biology
Developmental Biology
![]() Neuroscience
Neuroscience
![]() Immunology and Infection
Immunology and Infection
![]() Disease Mechanism Research
Disease Mechanism Research
![]() Drug Discovery and Toxicology
Drug Discovery and Toxicology
![]() Stem Cell and Regenerative Medicine
Stem Cell and Regenerative Medicine
FAQ
Q: Why is the requirement for cell viability so high after the preparation of single-cell suspension?
A: RNA from dead cells will be released into the extracellular fluid or mixed into the GEMs of living cells. These free RNAs may be encapsulated together with the cells, affecting subsequent reactions and ultimately leading to inaccurate analysis results. Meanwhile, a high number of dead cells will also lead to inaccurate estimation of cell quantity and affect the capture rate.
Q: How many cells can be sequenced in one run of 3' single-cell transcriptome sequencing? What is the capture efficiency of a single cell? Is there a possibility of capturing multiple cells?
A: Different platforms are equipped with microfluidic chips with 1–8 channels. Taking 10X as an example, a single chip can sequence up to 8 samples simultaneously. Each channel can process 100–10,000 cells, so a single chip can obtain 100–80,000 cells at one time.Under general experimental conditions, the single-cell capture efficiency of 3' single-cell transcriptome sequencing is as high as 65%. The probability of capturing multiple cells is extremely low, about 0.9% per 1000 cells.
Q: What results can be obtained from the standard analysis of scRNA sequencing?
A: The standard analysis pipeline includes quality control of sequencing data and statistics of various indicators (such as cell number, number of detected genes, data volume per single cell, etc.), gene expression quantification, cell type annotation, screening of differentially expressed genes and their functional annotation. In addition, personalized analysis can be performed based on the project background and experimental design.
Q: What common tissue types require nuclear extraction for single-nucleus transcriptome sequencing?
A: Brain tissues containing neurons, liver tissues targeting hepatocytes, lung tissues focusing on lung epithelial cells, adipose tissues for adipocytes, heart tissues for cardiomyocytes, muscle tissues for muscle cells, spinal cord/brain tissues targeting neurons, and kidney tissues focusing on podocytes, etc. At present, plant samples also adopt nuclear extraction for single-cell transcriptome research, which reduces the impact of stress gene expression and the interference of cytoplasmic metabolites, and is compatible with cells of larger diameters.
Q: Can fresh tissues be frozen after being placed in preservation solution for single-nucleus transcriptome sequencing?
A: No. This operation will form ice crystals and damage cells and cell nuclei.
