
- Home
- About Us
- Service
- Application
- Technology
- Resource
- Contact Us

circRNA Sequencing
Circular RNA (circRNA) sequencing is a precise method that identifies and quantifies stable, loop-structured RNAs, revealing their roles in gene regulation and potential as disease biomarkers. This technology is pivotal for advancing research in areas like cancer and neurodegenerative diseases, aiding in the development of new diagnostic and therapeutic approaches.
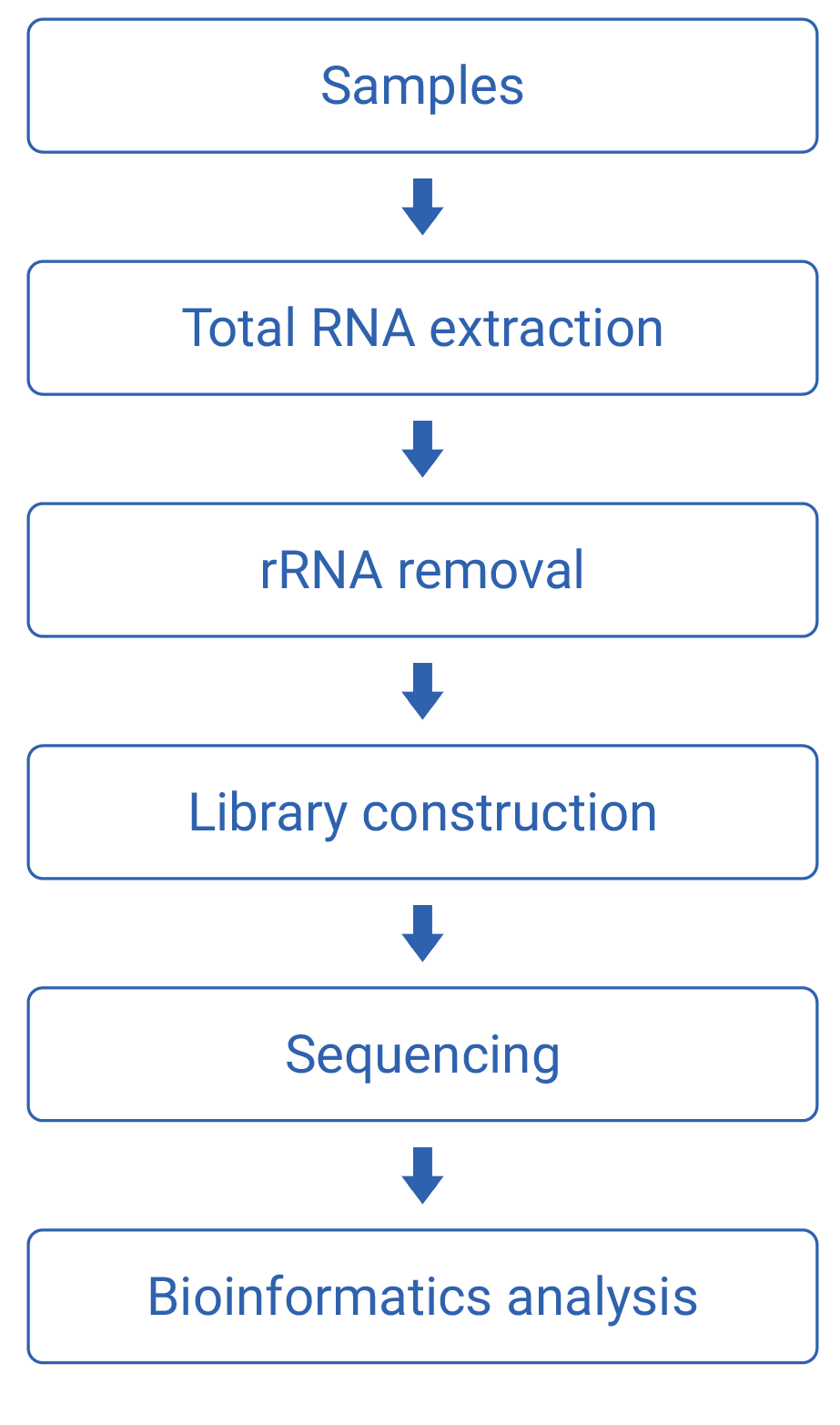
Work flow
Technical Parameters
| Sequencing range | mRNA+lncRNA+circRNA |
| Sequencing strategy | NGS PE150 |
| Sequencing throughput | 15Gb |
| Data quality | Fastq files, Q30≥85% |
| Data analysis | Standard analysis |
| TAT | Standard: 35 WD |
Applications
| Biomarker Discovery | Neuroscience | Aging Research |
| Cancer Research | Regulatory Functions | Cardiovascular Diseases |
Bioinformatics Analysis
1.Data Quality Control: Remove adapter contamination and low-quality reads from the raw data.
2.Reference Genome Alignment: Statistics of alignment results, display of alignment region distribution, and distribution of reads on chromosomes.
3.circRNA Prediction and Filtering: Results of circRNA prediction and filtering, statistical distribution of circRNA lengths, distribution of circRNA origins, and circRNA chromosomal distribution.
4.circRNA Expression Level Analysis: Statistics of circRNA expression levels, TPM density distribution, sample correlation, and principal component analysis.
5.circRNA Differential Expression Analysis: Results of circRNA differential expression, circRNA expression volcano plots, clustering of differentially expressed circRNAs, and Venn diagrams of differentially expressed circRNAs.
6.Analysis of circRNA Source Genes: GO, KEGG, Reactome functional enrichment analysis.
7.miRNA-circRNA Regulatory Network Analysis (customer must provide miRNA data from the same batch).
Advantages
![]() High Sensitivity
High Sensitivity
![]() High Specificity
High Specificity
![]() Scalability and Flexibility
Scalability and Flexibility
