- Home
- About Us
- Service
- Application
- Technology
- Resource
- Contact Us

LIUDUS®
HaploX's proprietary molecular technologies cover all the major steps within the wet lab NGS workflow, optimizing each step to ensure superior nucleic acid isolation and sequencing library preparation while significantly reducing sequencing errors. This comprehensive approach yields high-quality results with maximized efficiency.
01
CUBE-ctDNA
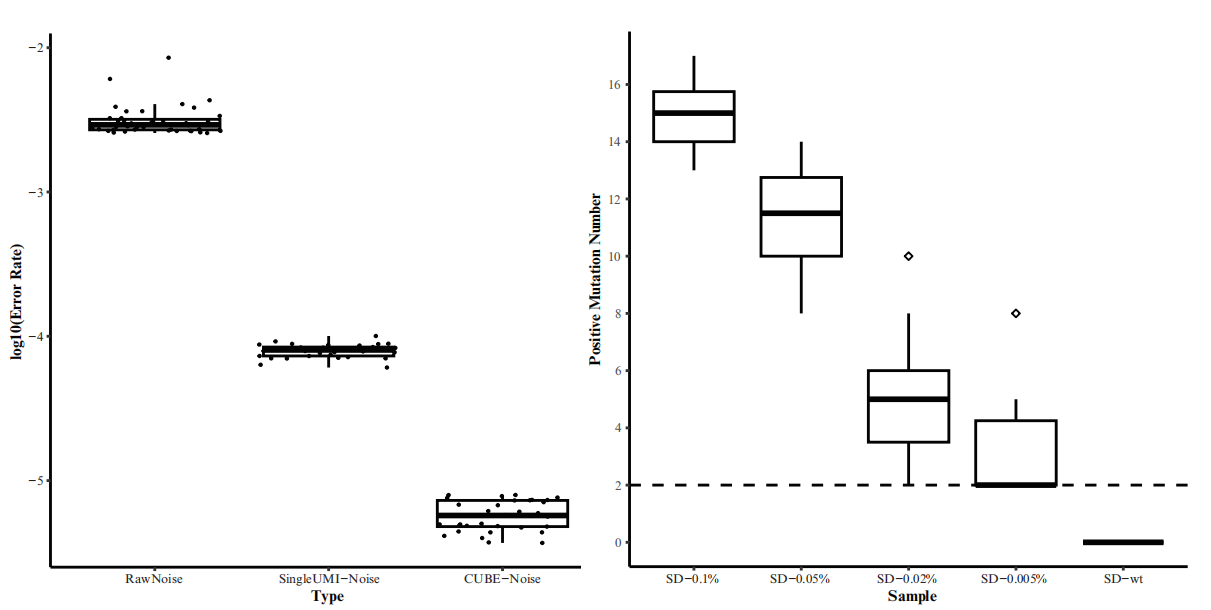
CUBE-ctDNA is HaploX's innovative single-molecule barcoding technology designed to enhance the signal-to-noise ratio in ultra-deep sequencing. During library preparation, this technology assigns two unique molecular identifiers (UMIs) to both ends of each double-stranded DNA molecule. This allows each sequence read, post-PCR amplification and sequencing, to be traced back to the original DNA molecule. True mutations appear consistently in reads tagged with the same UMIs, while false mutations, such as PCR and sequencing errors, appear only in a fraction of these reads. By attaching UMIs to both DNA strands, CUBE-ctDNA leverages sequence complementarity to filter out false mutations detected in only one strand.
Through CUBE-ctDNA technology, we significantly reduce NGS background noise from an industry standard of 0.1% to 0.00058% in preclinical validation studies, allowing ultra-deep sequencing with a ctDNA LoD as low as 0.005%.
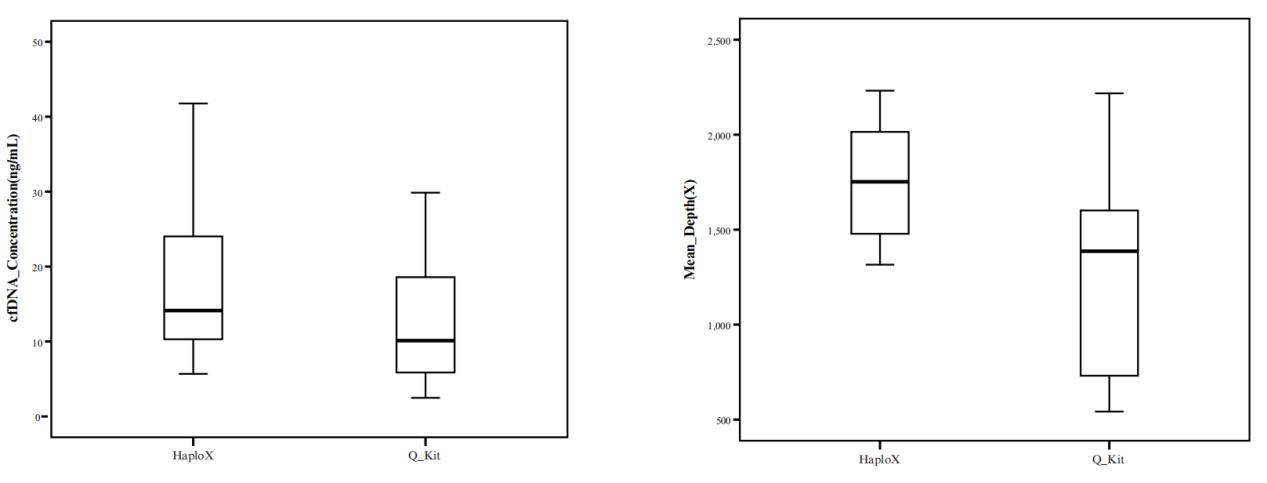
Compared to a competitive international DNA extraction product in our preclinical validation studies, Effentration® consistently achieved higher cfDNA isolation efficiency and greater average sequencing depth.
02
Effentration®
DNA extraction is a critical first step to the success of NGS-based variant detection. To that end, HaploX developed Effentration® for effective cfDNA extraction. Via the use of lysis, binding and washing solutions with proprietary formulations, Effentration® can effectively enrich low-abundance cfDNA and remove undesirable nucleic acid contaminants.
03
HAPCap®
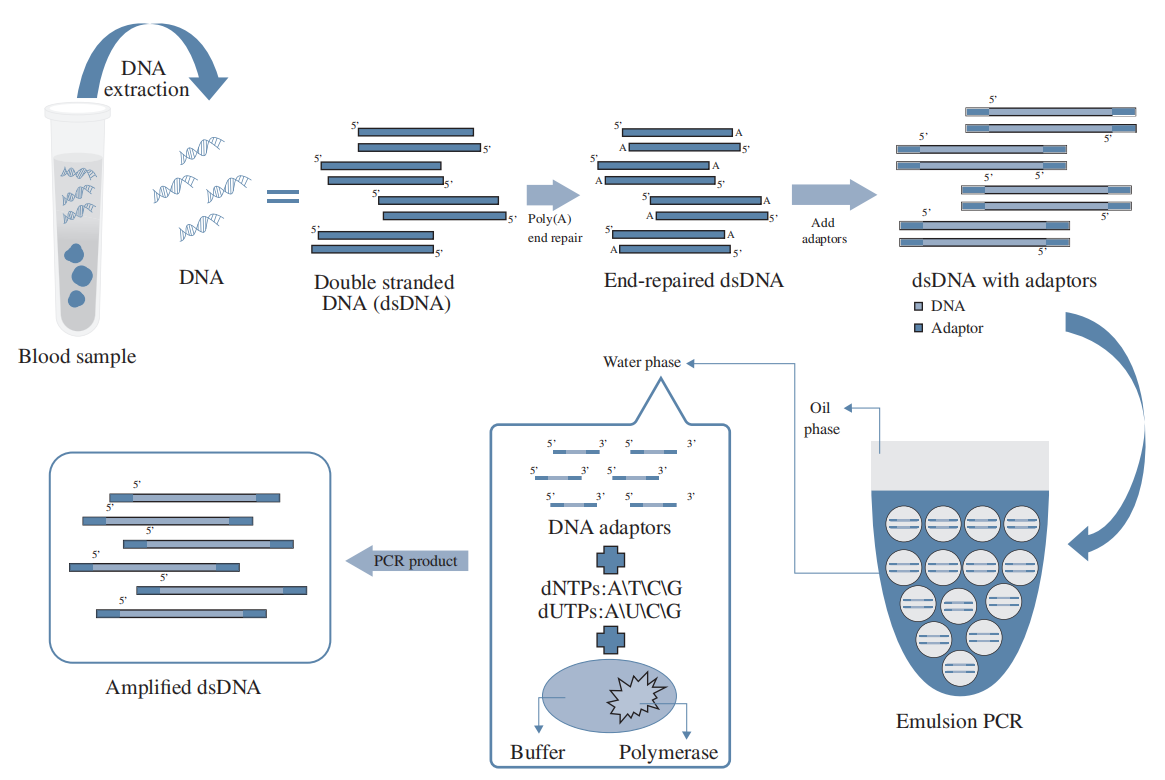
During library preparation, the initial DNA material from biopsy samples needs PCR amplification to ensure sufficient DNA for sequencing. Since ctDNA is a minor fraction of cfDNA, conventional PCR amplification dilutes detection probability by favoring dominant cfDNA types. To tackle this, HaploX developed HAPCap® for ctDNA enrichment during DNA capture. This process begins with emulsion PCR, where input DNA molecules are amplified in separated micro-volume water-in-oil droplets. Then, ctDNA-specific, biotin-labeled DNA probes hybridize to ctDNA, which is captured using streptavidin magnetic beads. HAPCap® effectively removes PCR amplification bias, enabling selective amplification of low-abundance ctDNA to enhance mutation detection sensitivity.
HAPCap® Technical Flow Chart